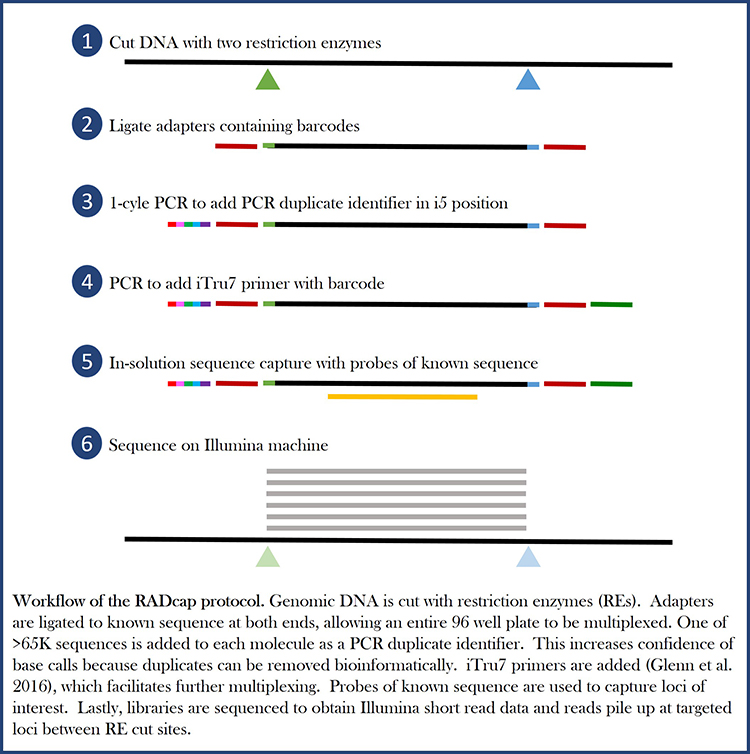
RADcap is a further reduced representation library preparation method that combines RADseq and sequence capture. Like RADseq, RADcap is cheap to do and no prior genetic information from the species is needed. Like sequence capture, RADcap controls which loci will be recovered in the resulting dataset, and there is little missing data. RADcap makes an advantage on both of these methods by allowing the researchers to bioinformatically remove duplicate reads that resulted from PCR. These duplicate reads are not only unnecessary, but lead to false confidence in SNP calls.
Read the RADcap paper here.
Here is a clip from a seminar talking about how well RADcap worked on a previous project.
RADcap is based on 3RAD, a dual-digest RADseq variant that uses a third enzyme to break apart adapter dimers and undesired products during the ligation. This makes the reaction much more efficient, so less input DNA can be used. Watch the Adapterama III video to learn about 3RAD, the first steps of OrphanCap.
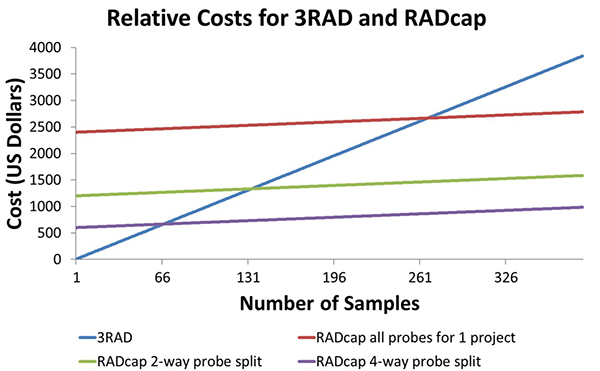
Here is a picture of the price comparison between RADcap and 3RAD. The relative price of 3RAD and RADcap is dependent on the number of samples in the study. The blue line is 3RAD, and the cost increases by $13 per sample. The red line is RADcap. This shows that with around 260 samples- 3 plates, RADcap becomes more economical than 3RAD, and it is worth it to invest in buying the probes. It is also possible to combine probe sets for multiple projects. If two projects are combined, it is as if the cost of sequence capture is cut in half, and only 131 samples make RADcap cheaper. If 4 projects are combined into a single probe set, only 66 samples are needed to make RADcap cheaper than 3RAD.
Check out this Google Spreadsheet to find scientists that want to share baitsets.
3RAD is based on iTru primers, developed by Travis Glenn at the University of Georgia and Brant Faircloth at Louisiana State University, which allow multiplexing of thousands of samples and sequencing on Illumina machines.
Watch the Adapterama I video to learn about how we can multiplex so many samples.
OrphanCap is funded by the National Science Foundation Postdoctoral Research Fellowship in Biology under Grant No. 1711794. RADcap was developed by Sandra L. Hoffberg, Troy J. Kieran, Julian M. Catchen, Alison Devault, Brant C. Faircloth, Rodney Mauricio, and Travis C. Glenn. RADcap was supported by grants DEB-1242260 and DEB-1146440 from the U.S. National Science Foundation and the U.S. National Science Foundation Partnership for International Research and Education (PIRE) program (OISE 0730218). 3RAD and Adapterama I were developed by Travis C. Glenn, Roger Nilsen, Troy J. Kieran, John W. Finger, Todd W. Pierson, Kerin E. Bentley, Sandra L. Hoffberg, Swarnali Louha, Francisco J. Garcia-De-Leon, Miguel Angel del Rio Portilla, Kurt Reed, Jennifer L. Anderson, Jennifer K. Meece, Sammy Aggery, Romdhane Rekaya, Magdy Alabady, Myriam Belanger, Kevin Winker, and Brant C. Faircloth. That work was partially supported by DEB-1242241, DEB-1242260, DEB-596 1136626, DEB-1146440, DGE-0903734, and OISE 0730218 from the U.S. National Science 597 Foundation, SAGARPA-FIRCO (grant RGA-BCS-12-000003), UCMEXUS (grant CN-13-617), 598 and CB-CONACYT (grant 157993). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the NSF.
© Copyright 2018, Sandra L. Hoffberg, Ph.D.